What is SBGN?
The Systems Biology Graphical Notation (SBGN) is a standard used to represent molecular networks graphically.
It includes three languages: Process Description (PD), that allows representing modulated processes such as catalyzed reactions; Activity Flow (AF), that may be used to represent biomolecular activities and the influences they have on each other; and Entity Relationship (ER), that permits representing relationships between biomolecular entities such as molecular interactions.
More information can be found on the SBGN website.
What is an SBGN brick?
An SBGN brick is a graphical representation of a biomolecular or biological concept commonly found in molecular networks, expressed using one of the three SBGN languages (PD, AF, or ER).
We distinguish two types of bricks: template bricks and instance bricks.
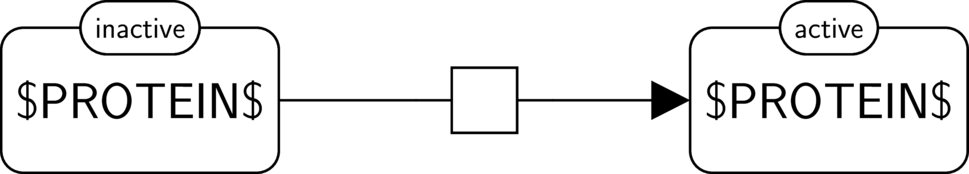
A template brick is a pattern representing a certain kind of biomolecular process, activity or influence (e.g. a protein phosphorylation, a protein kinase activity, a positive influence of a protein kinase activity on another protein kinase activity).
An example of a template brick is given if the figure below, panel A.
An instance brick is a representation of a specific instance of a concept represented by a template brick (e.g. the phosphorylation of the RSK protein, the protein kinase activity of the RSK protein, the positive influence of the protein kinase acitivity of ERK on the protein kinase acitivty of RSK).
Two examples of an instance brick are given in the figure below, panel B.

How are they useful?
Bricks may be used as a teaching aid for those learning SBGN or systems biology.
Template bricks may also be used to generate specific instance bricks (template-based construction) or to match real instance bricks found in SBGN maps (annotation).
How are they associated to concepts?
SBGN bricks represent biomolecular or biological concepts.
To formally associate the bricks with the concepts they represent, we build the BricKs Ontology (BKO).
BKO includes terms that describe concepts, the template bricks representing these concepts, and categories that gather bricks in a broader way.
BKO can be browsed on this page.
How are they represented?
SBGN Bricks are represented using the three SBGN languages.
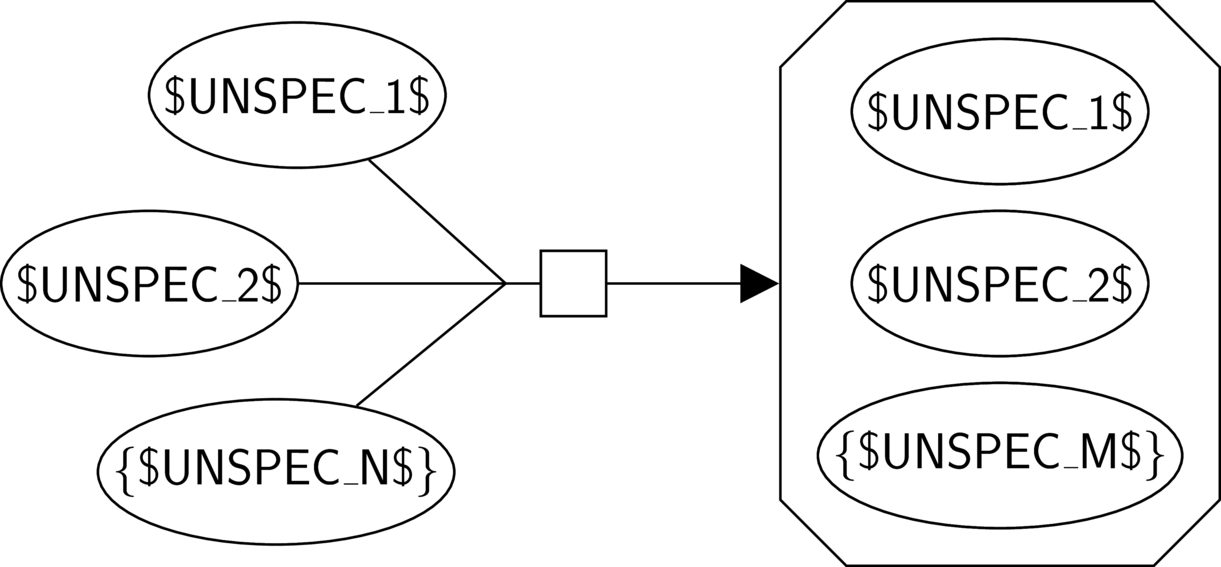
Template bricks are also represented using additional textual constructs, that allow expressing generality or repetition, for example.
The main constructs are the following:
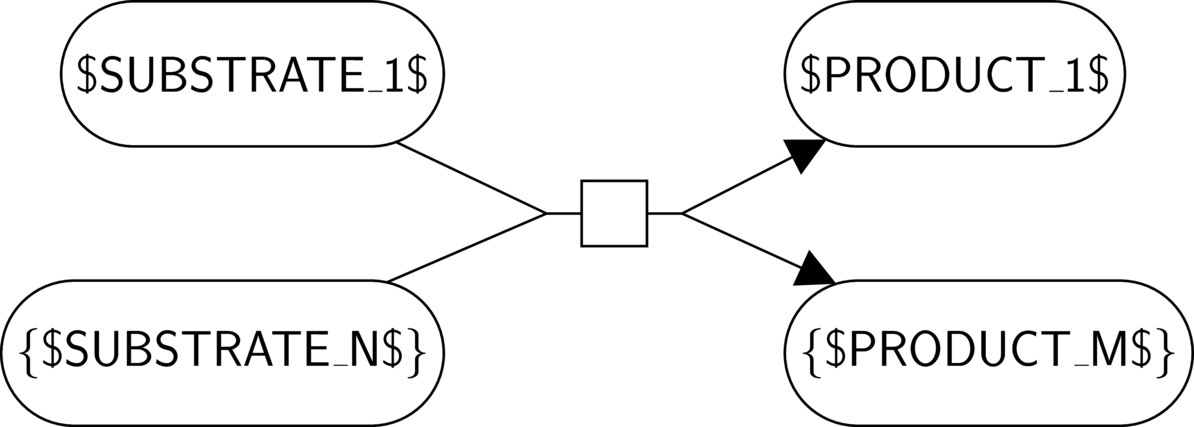
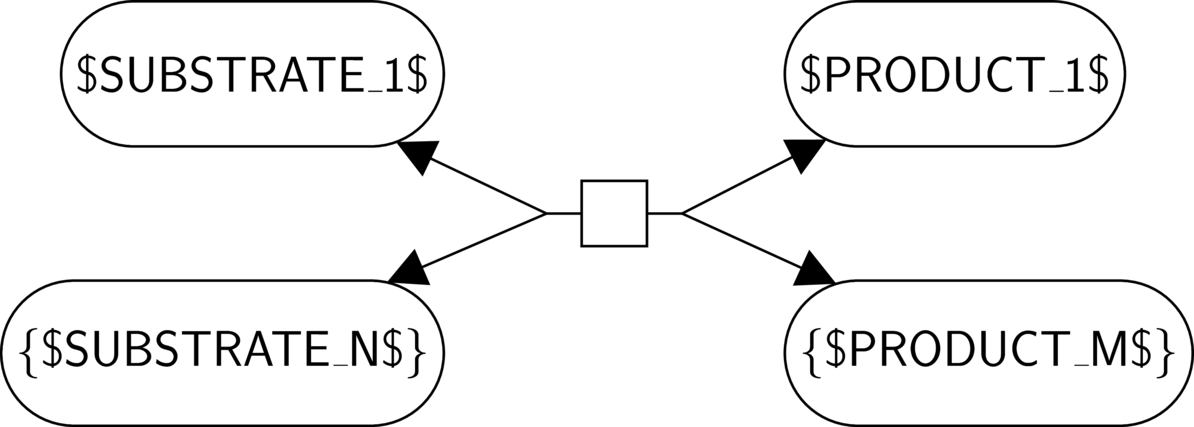
Literal: any literal not enclosed by reserved characters ("$", "[" and "]", "{" and "}", and "!") represents itself.
Variable string: $NAME$ represents any non-empty string. The string is named by "NAME", and every occurrence of $NAME$ represents the same string.
Optional string: [$NAME$] represents any string, or the empty string.
Repetition: {$NAME$} applies to the whole glyph and represents a repetition of this glyph of zero or more times.
Absence: !$NAME$! applies to the whole glyph and represents the absence of a glyph.
More examples?
The following alignment gives a list of common PD template bricks (in blue) and their corresponding AF (in yellow) and ER (in pink) template bricks.
The alignment is organized following broad categories.
Note that processes such as metabolic reactions cannot be correctly represented in AF.
However if one absolutely needs such a representation, one may refer to link for examples.
BKO:0000571 complex association and dissociation
|

|

|
|
BKO:0000204 non-covalent binding (PD narrow 1)
|
BKO:0000203 binding (AF main)
BKO:0000042 combined necessary stimulation of binding activities on another biological activity (AF main)
|
BKO:0000207 non-covalent binding (ER main) Should be broad
|
In the PD part, we assume that the label of the complex is $COMPLEX$. In the AF part, we show a number of binding activities, and their possible combined necessary stimulation on another biological activity, here the one of $COMPLEX$. We consider the case where $COMPLEX$ indeed has an activity, and thus where the binding activities of its components are necessary for this activity to take place.
BKO:0000221 protein multimerisation

|

|

|
|
BKO:0000223 protein multimerisation (PD main)
|
BKO:0000203 binding (AF main)
BKO:0000059 simple necessary stimulation of a binding activity on another biological activity (AF main)
|
BKO:0000222 protein multimerisation (ER main)
|
In the AF part, $COMPLEX$ is a new label, that can be built from the label $PROTEIN$ for example. In this part, we show the binding activity of $PROTEIN$ and its possible necessary stimulation on another biological activity, here the one of $COMPLEX$. We consider the case where $COMPLEX$ indeed has an activity, and thus where the activity of its unique component is necessary for this activity to take place.

|
Not applicable
|
Not applicable
|
|
BKO:0000170 dissociation (PD narrow 1)
|
|
|
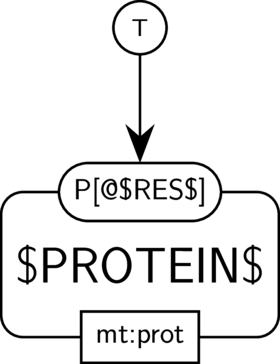
BKO:0000574 phosphorylation and dephosphorylation
BKO:0000438 protein phosphorylation

|
Not applicable
|
|
|
BKO:0000440 protein phosphorylation (PD narrow 1)
|
|
BKO:0000439 protein phosphorylation (ER main)
|
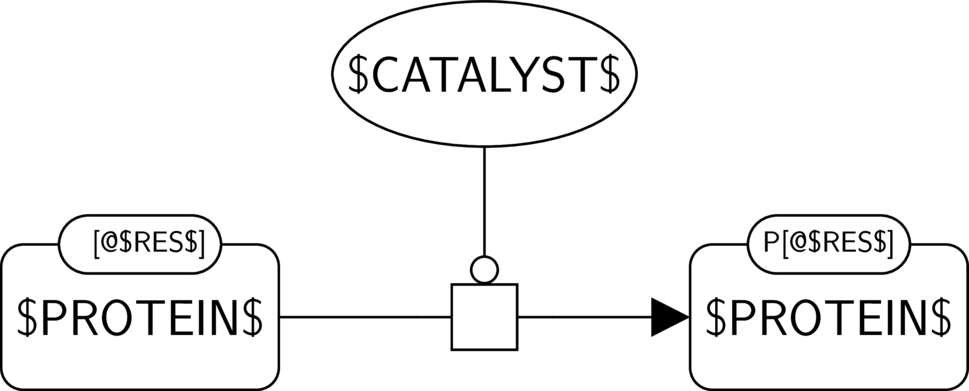
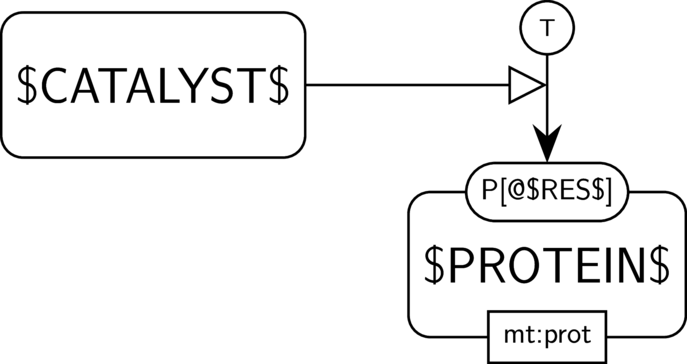
GO:0004672 protein kinase activity
|

|
|
|
BKO:0000287 protein kinase activity (PD narrow 1)
|
BKO:0000285 protein kinase activity (AF main)
BKO:0000071 simple positive influence of a protein kinase activity on another biological activity (AF main)
|
BKO:0000282 protein kinase activity (ER broad 1)
|
In the AF part, we show the protein kinase activity of $CATALYST$ and its possible influence on another biological activity, here the one of $PROTEIN$. We assume that $PROTEIN$ indeed has an activity, and that its phosphorylated form is its active form, which is often the case in signalling pathways.
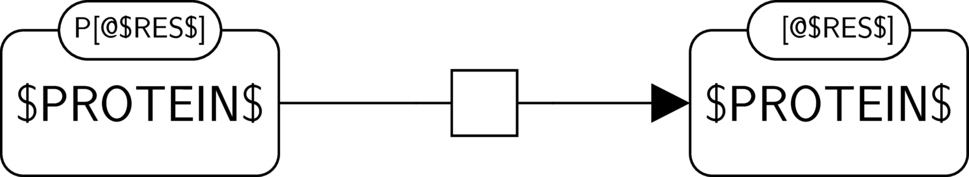
BKO:0000375 protein dephosphorylation
|
Not applicable
|
|
|
BKO:0000370 protein dephosphorylation (PD narrow 1)
|
|
BKO:0000369 protein dephosphorylation (ER main)
|
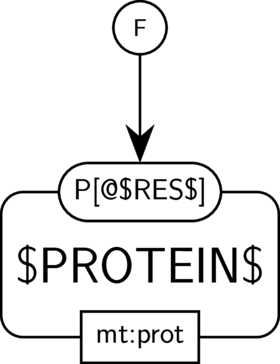
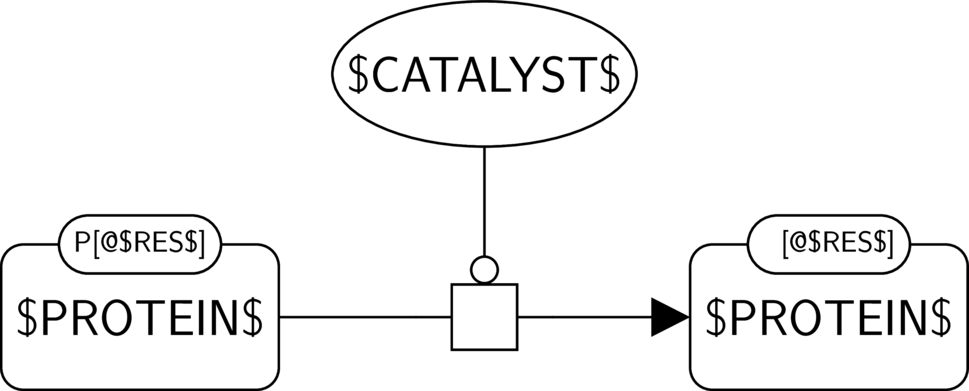
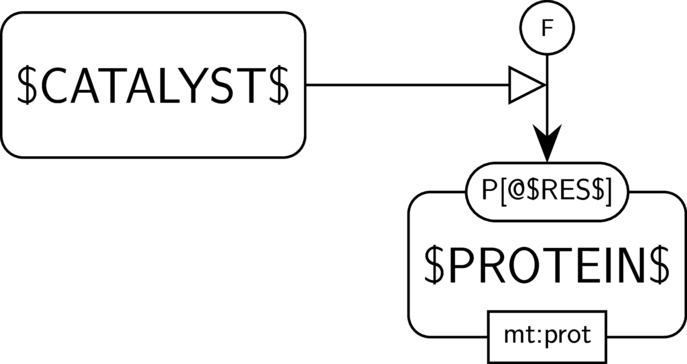
GO:0004721 phosphoprotein phosphatase activity
|

|
|
|
BKO:0000233 phosphoprotein phosphatase activity (PD narrow 1)
|
BKO:0000068 simple negative influence of a phosphoprotein phosphatase activity on another biological activity (AF main)
|
BKO:0000228 phosphoprotein phosphatase activity (ER broad 1)
|
In the AF part, we show the phosphoprotein phosphatase activity of $CATALYST$ and its possible influence on another biological activity, here the one of $PROTEIN$. We assume that $PROTEIN$ indeed has an activity, and that its phosphorylated form is its active form, which is often the case in signaling pathways.
In a similar way other types of addition and removal of chemical groups may be represented:
|
Not applicable
|
|
|
BKO:0000498 activation (PD narrow 1)
|
|
BKO:0000495 activation (ER main)
|
BKO:0000208 deactivation

|
Not applicable
|

|
|
BKO:0000211 deactivation (PD narrow 1)
|
|
BKO:0000210 deactivation (ER main)
|
BKO:0000599 metabolism
BKO:0000585 metabolic reaction
BKO:0000511 irreversible metabolic reaction
|
Not applicable
|
Not applicable
|
|
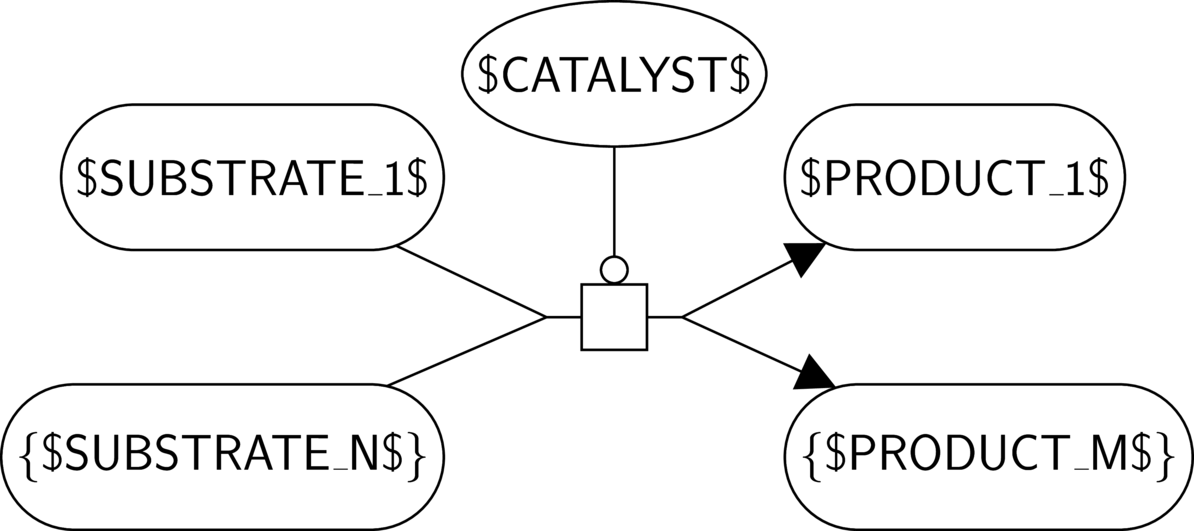
BKO:0000513 irreversible metabolic reaction (PD main)
|
|
|
BKO:0000514 reversible metabolic reaction
|
Not applicable
|
Not applicable
|
|
BKO:0000515 reversible metabolic reaction (PD main)
|
|
|
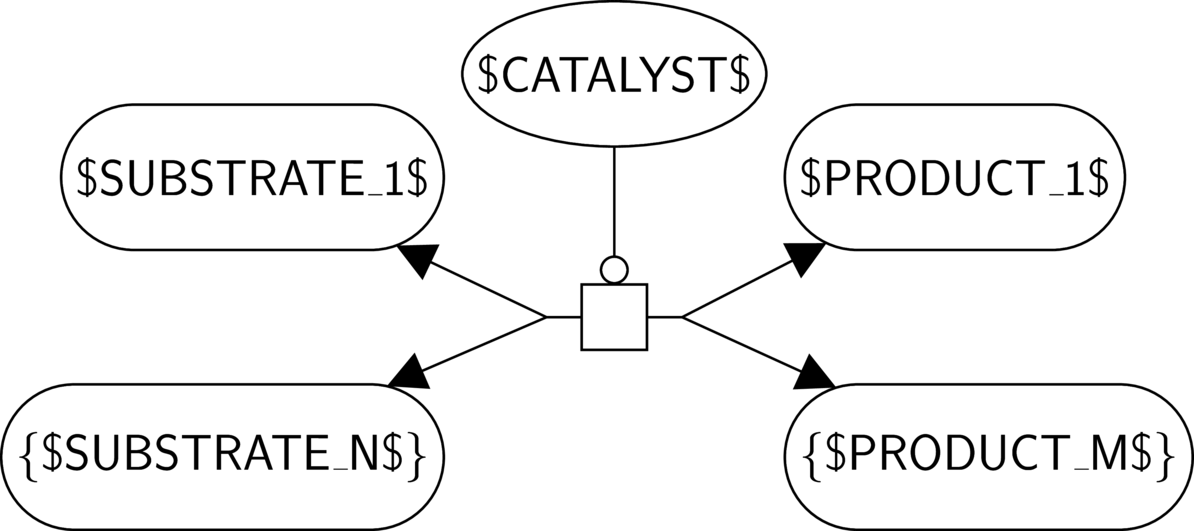
BKO:0000196 metabolic catalytic activity
|
Not applicable
|
Not applicable
|
|
BKO:0000197 metabolic catalytic activity (PD narrow 1)
|
|
|
|
Not applicable
|
Not applicable
|
|
BKO:0000198 metabolic catalytic activity (PD narrow 2)
|
|
|
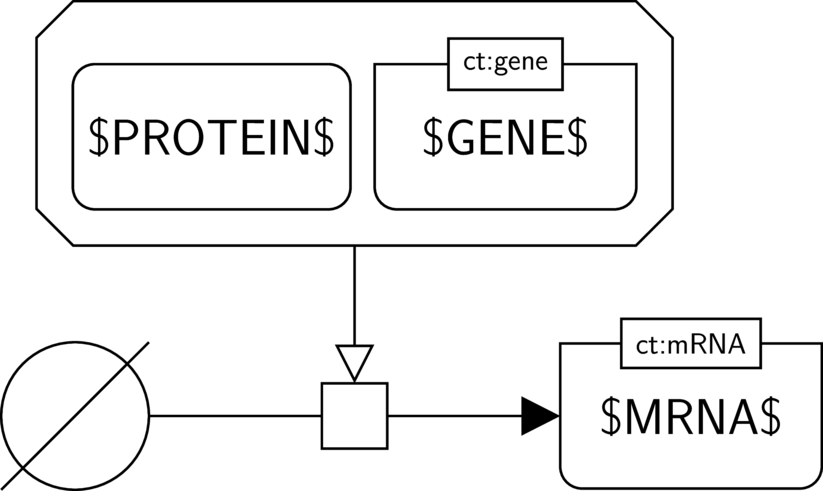
BKO:0000597 gene expression
BKO:0000573 transcription
SBO:0000183 transcription
|

|
|
|
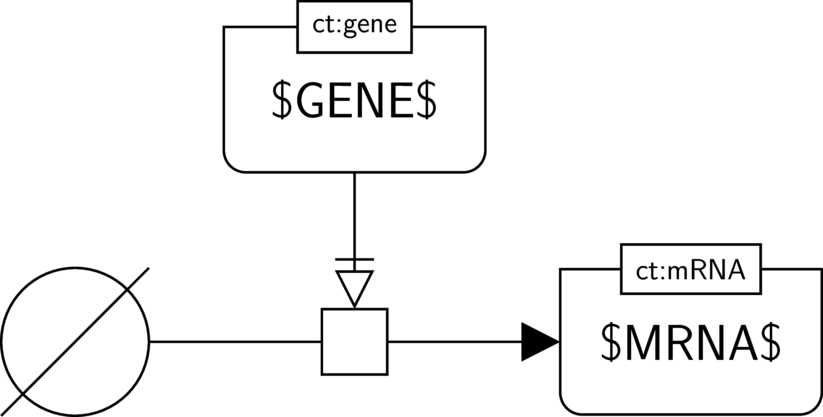
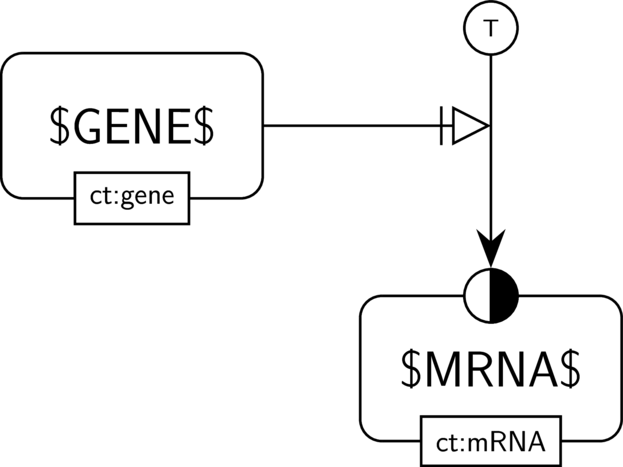
BKO:0000477 transcription (PD main)
|
BKO:0000061 simple necessary stimulation of a biological activity on another biological activity (AF main)
|
BKO:0000478 transcription (ER main)
|
In the AF part we show the biological activity of $GENE$ and its necessary stimulation on another biological activity, here the one of $MRNA$. We assume that $MRNA$ indeed has an activity (the one of guiding translation). Hence the activity of the gene (the one of guiding transcription) is necessary for this activity to take place.
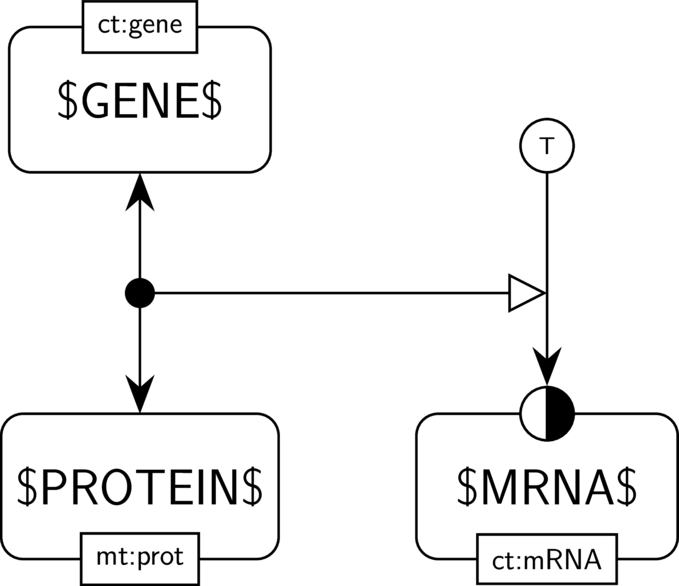
GO:0001216 DNA-binding transcription activator activity
|

|
|
|
BKO:0000518 DNA-binding transcription activator activity (PD main)
|
BKO:0000520 DNA-binding transcription activator activity (AF main)
BKO:0000055 combined positive influence of a DNA-binding transcription activator activity and a biological activity on another biological activity (AF main)
|
BKO:0000519 DNA-binding transcription activator activity (ER main)
|
In the AF part we show the DNA-binding transcription activator activity of $PROTEIN$, the biological activity of $GENE$, and their combined positive influence on another biological activity, here the one of $MRNA$. We assume that $MRNA$ has an activity (the one of guiding translation). Hence the activity of $PROTEIN$ and the activity of $GENE$ (the one of guiding transcription) together have a positive influence on the activity of $MRNA$. We do not use a necessary stimulation because other transcription factors may be sufficient to enhance the translation of $GENE$, and thus the occurrence of the activity of $PROTEIN$ might not be necessary for the one of $MRNA$ to take place.
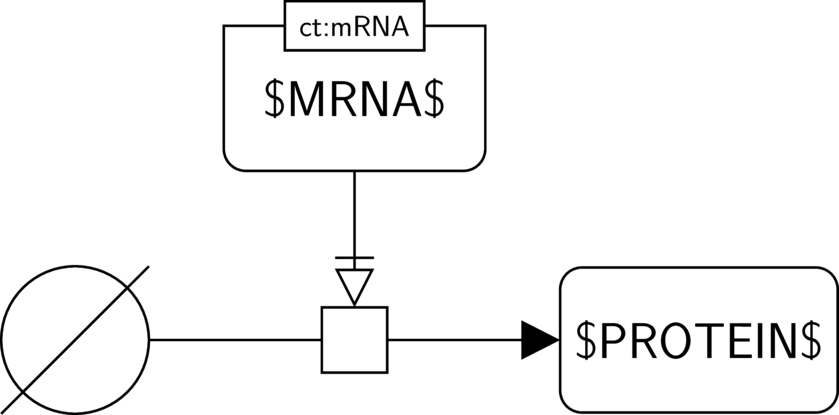
BKO:0000589 translation
SBO:0000184 translation
|

|
|
|
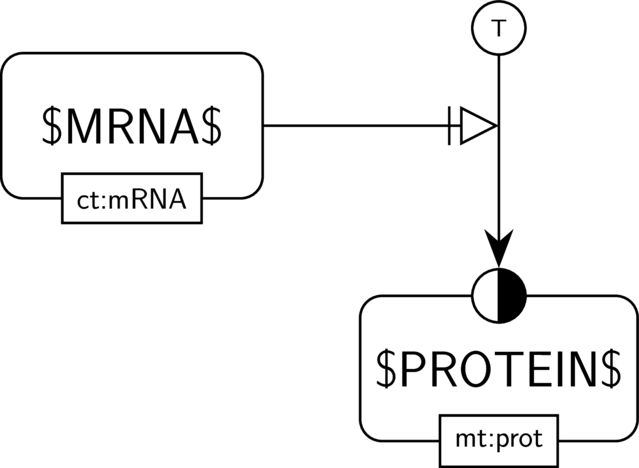
BKO:0000481 translation (PD main)
|
BKO:0000061 simple necessary stimulation of a biological activity on another biological activity (AF main)
|
BKO:0000480 translation (ER main)
|
In the AF part we show the biological activity of $MRNA$ and its possible necessary stimulation on another biological activity, here the one of $PROTEIN$. We consider the case where $PROTEIN$ indeed has an activity, and thus where the activity of $MRNA$ is necessary for this activity to take place.
BKO:0000598 translocation and transport
BKO:0000594 channel opening and closing
BKO:0000508 channel opening
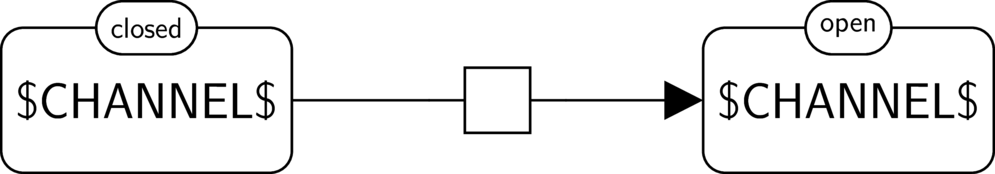
|
Not applicable
|
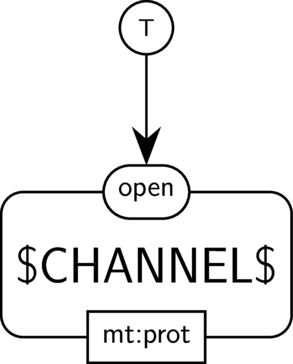
|
|
BKO:0000509 channel opening (PD main)
|
|
BKO:0000510 channel opening (ER main)
|
BKO:0000505 channel closing
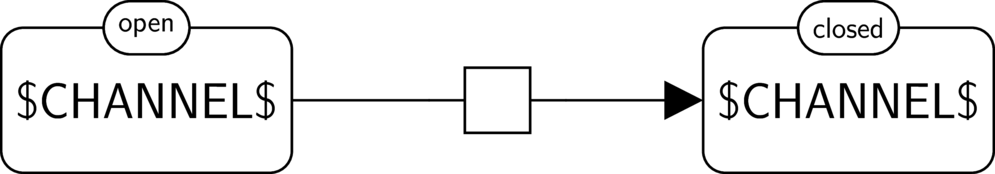
|
Not applicable
|
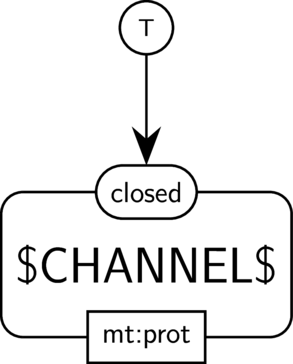
|
|
BKO:0000506 channel closing (PD main)
|
|
BKO:0000507 channel closing (ER main)
|
BKO:0000590 translocation
SBO:0000185 translocation reaction

|
Not applicable
|

|
|
BKO:0000484 translocation reaction (PD narrow 2)
|
|
BKO:0000482 translocation reaction (ER main)
|
BKO:0000575 transport
SBO:0000655 transport reaction - GO:0005215 transporter activity

|

|

|
|
BKO:0000090 transport reaction (PD narrow 2)
|
BKO:0000088 transporter activity (AF main)
BKO:0000073 simple positive influence of a transporter activity on another biological activity (AF main)
|
BKO:0000087 transport reaction (ER main)
|
In the AF part, we show the transport activity of $TRANSPORTER$ and its possible positive influence on another biological activity, the one of $UNSPEC$. We consider the case where $UNSPEC$ indeed has an activity when in compartment $COMP$.












































